Dynamics in Transcriptomics: Advancements in RNA-seq Time Course and Downstream Analysis - ScienceDirect

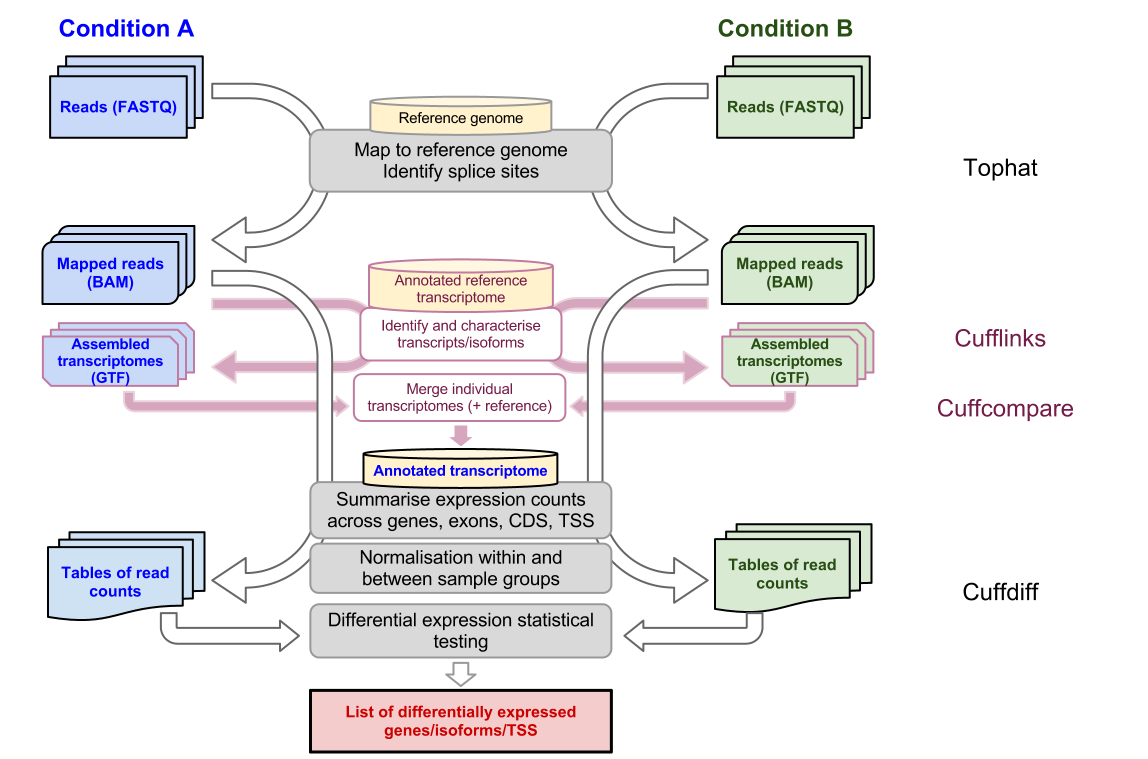
Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks | Nature Protocols
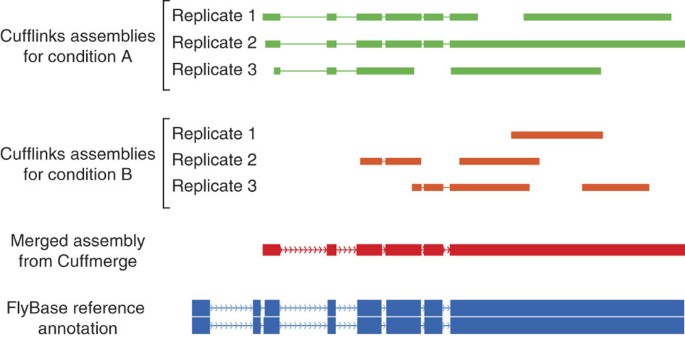
Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks | Nature Protocols

Biosensors | Free Full-Text | A Comparison of Methods for RNA-Seq Differential Expression Analysis and a New Empirical Bayes Approach

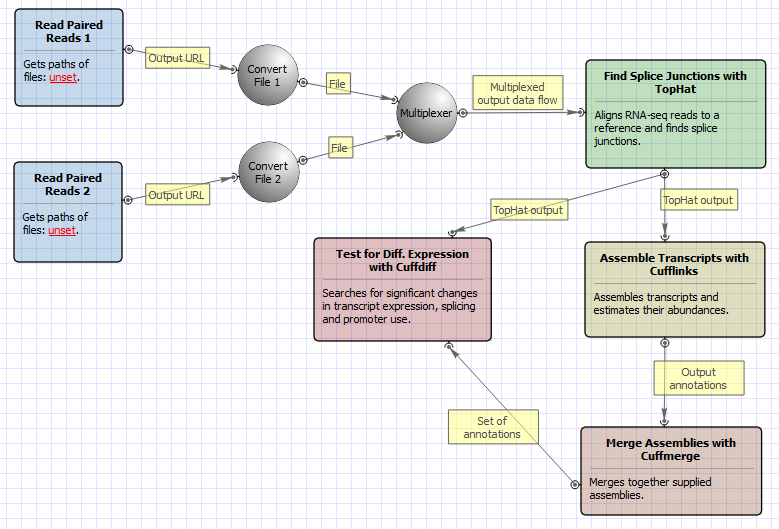
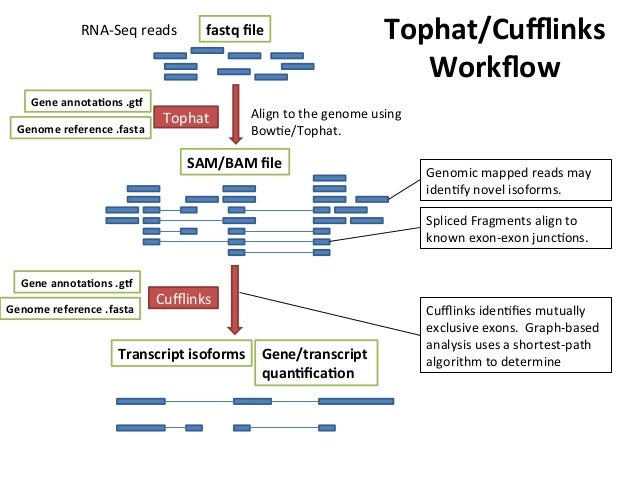
Introduction to tophat cufflinks workflow - Bioinformatics Team (BioITeam) at the University of Texas - UT Austin Wikis

Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation | Nature Biotechnology
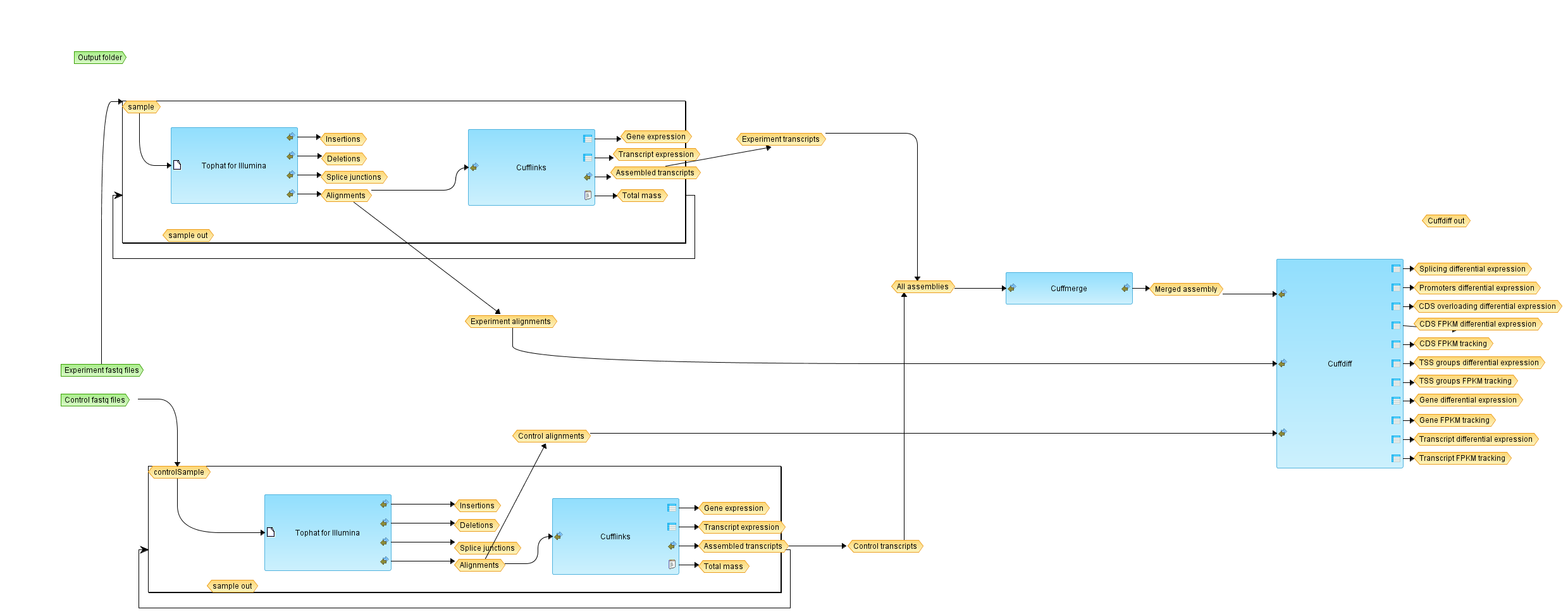
Quantification of RNA-seq with Cufflinks (with de-novo assembly) for FASTQ files (workflow) - BioUML platform

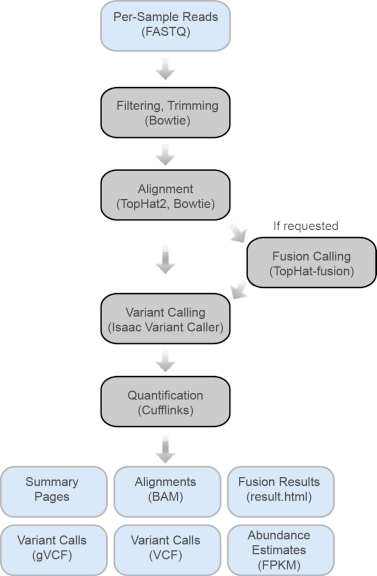
Example of the RNA-Seq analysis pipeline. (A) Typical simple pipeline.... | Download Scientific Diagram
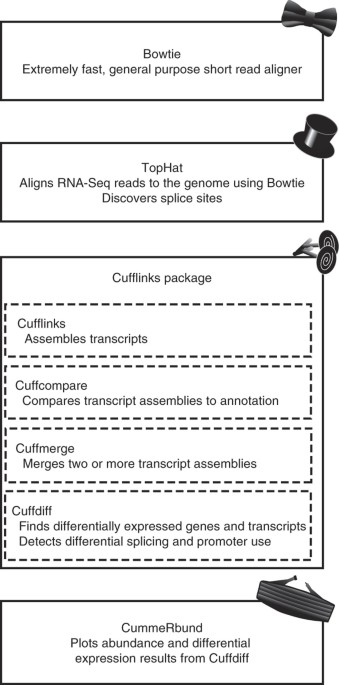
Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks | Nature Protocols
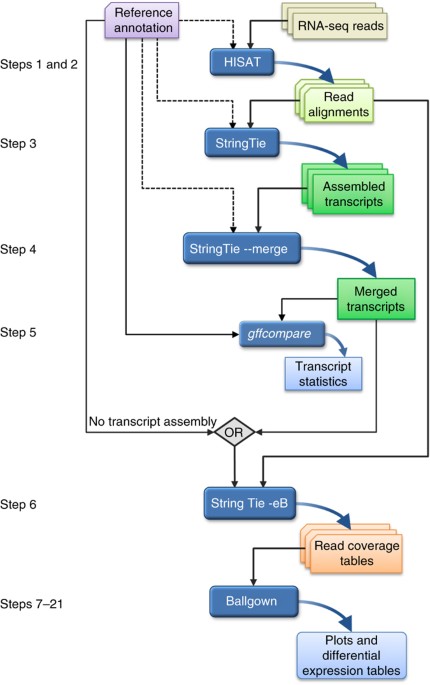
Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown | Nature Protocols